As is customary, eukaryotic promoters are more complex. Eukaryotes have three types of RNAPs, resulting in three corresponding classes of promoters. RNAP I is responsible for transcribing rRNA and corresponds to class I promoters, consisting of a core promoter (-45 to +20) and upstream control elements (-180 to -107).
Small RNAs have three types of class III promoters. The promoters for 5S rRNA and tRNA genes are downstream promoters, located downstream of the transcription start point. They contain box A, box B, and box C elements and require recognition by transcription factors such as TFIIIA and TFIIIC. The promoter for snRNA is located upstream of the transcription start point.
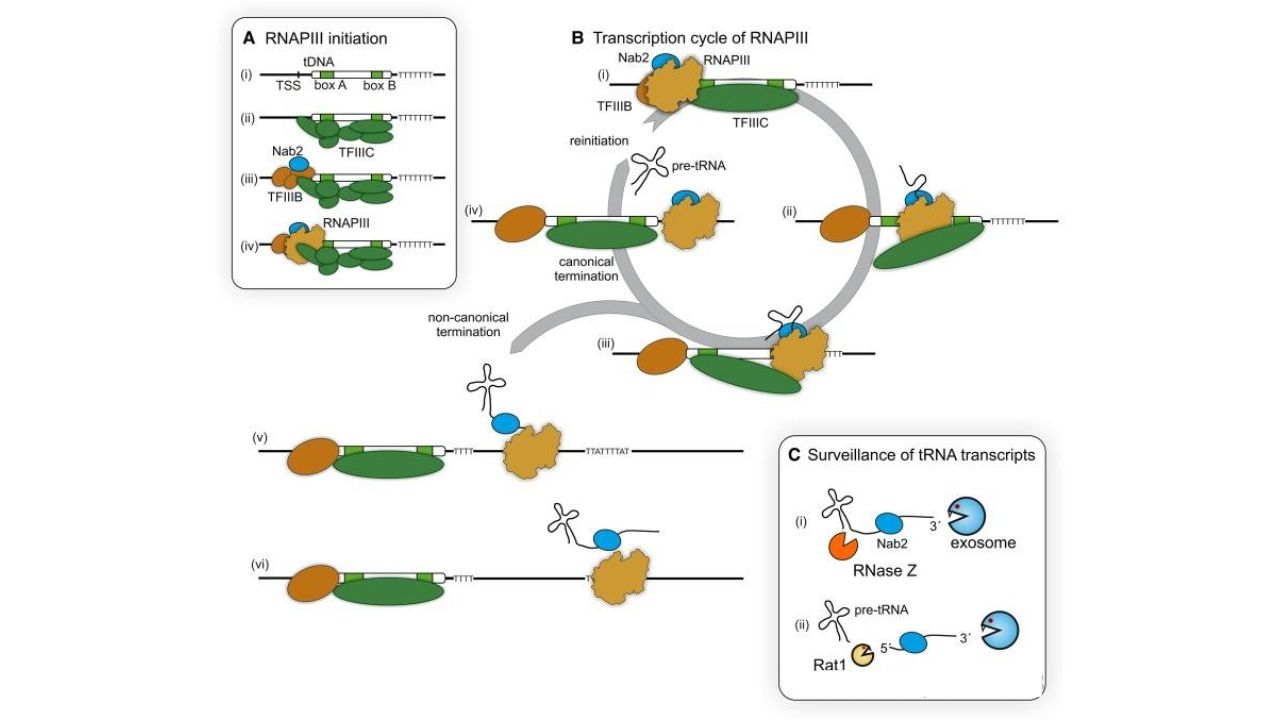 RNAP III promoters and transcription process. Biochem Soc Trans. 2016.
RNAP III promoters and transcription process. Biochem Soc Trans. 2016.
Class II promoters, corresponding to RNAP II (Pol II) for transcribing protein-coding genes, are the most studied. Its TATA box corresponds to the prokaryotic Pribnow box, located at positions -25 to -32; around position -80 is the CAAT box, corresponding to the prokaryotic -35 sequence; there are also GC boxes and octamer boxes, as well as some response elements.
The TATA box is also called the basal promoter. It, along with the upstream TFIB recognition element (BRE), the initiator (Inr) centered at the transcription start site, and the downstream promoter element (DPE), constitute the core promoter. All others are called upstream elements.
In eukaryotic transcriptional regulation, another commonly used cis element is the enhancer. Enhancers are DNA elements that bind to cofactors and transcription factors (TFs) and can increase the transcriptional level of their target genes by directly stimulating the promoter (usually through chromatin loops).
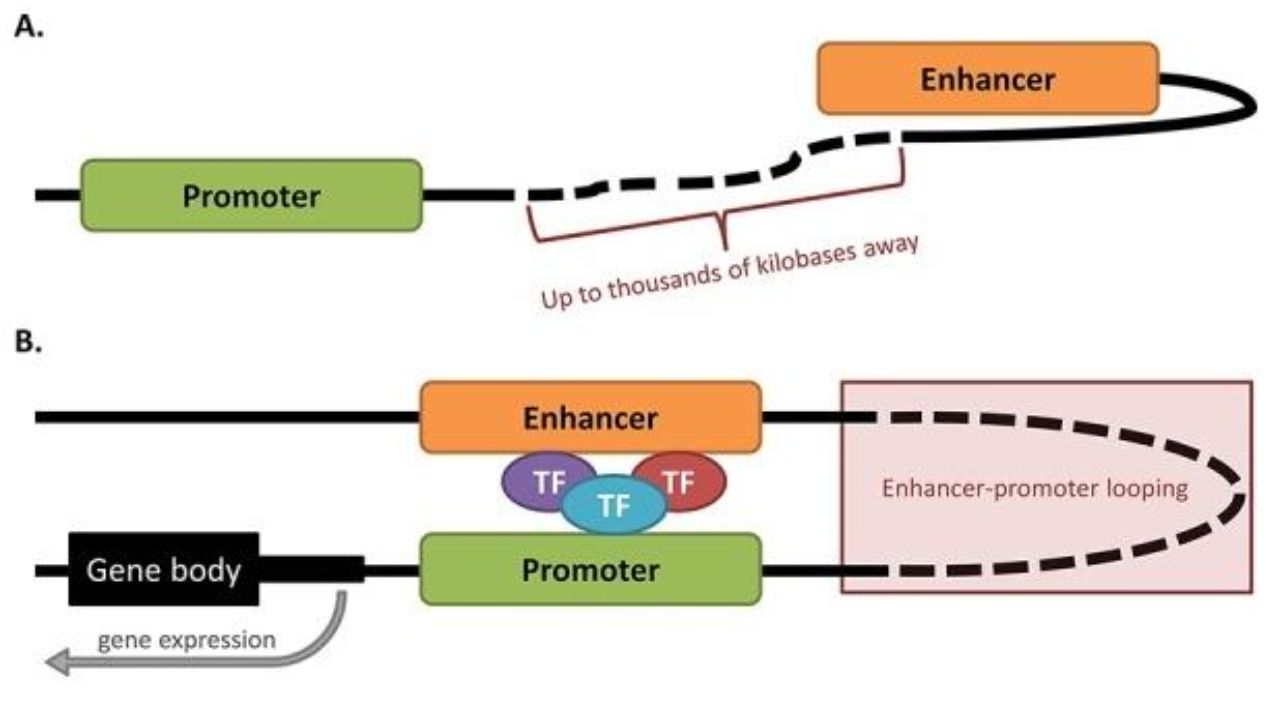 Enhancers regulate transcription through the promoter. Oncotarget. 2015.
Enhancers regulate transcription through the promoter. Oncotarget. 2015.
The location of a promoter is fixed, meaning it has a specific distance and orientation from the gene. Enhancers, however, are different; they may be thousands of kb away from the promoter they regulate, either upstream or downstream. In an inactive state, the two do not approach each other. But in an active state, enhancers approach the promoter in three-dimensional space by forming a loop structure, recruiting corresponding transcription factors to that region, thereby promoting gene expression.
Transcriptional regulation is closely related to chromatin structure. Active enhancers typically lack nucleosome structures to facilitate TF binding. Nearby histones often possess epigenetic markers, such as H3K4me1 (monomethylation of lysine 4 of histone H3) and acetylation of H3K27 (H3K27ac). Trimethylation of H3K4 (H3K4me3) is typically enriched on gene promoters.
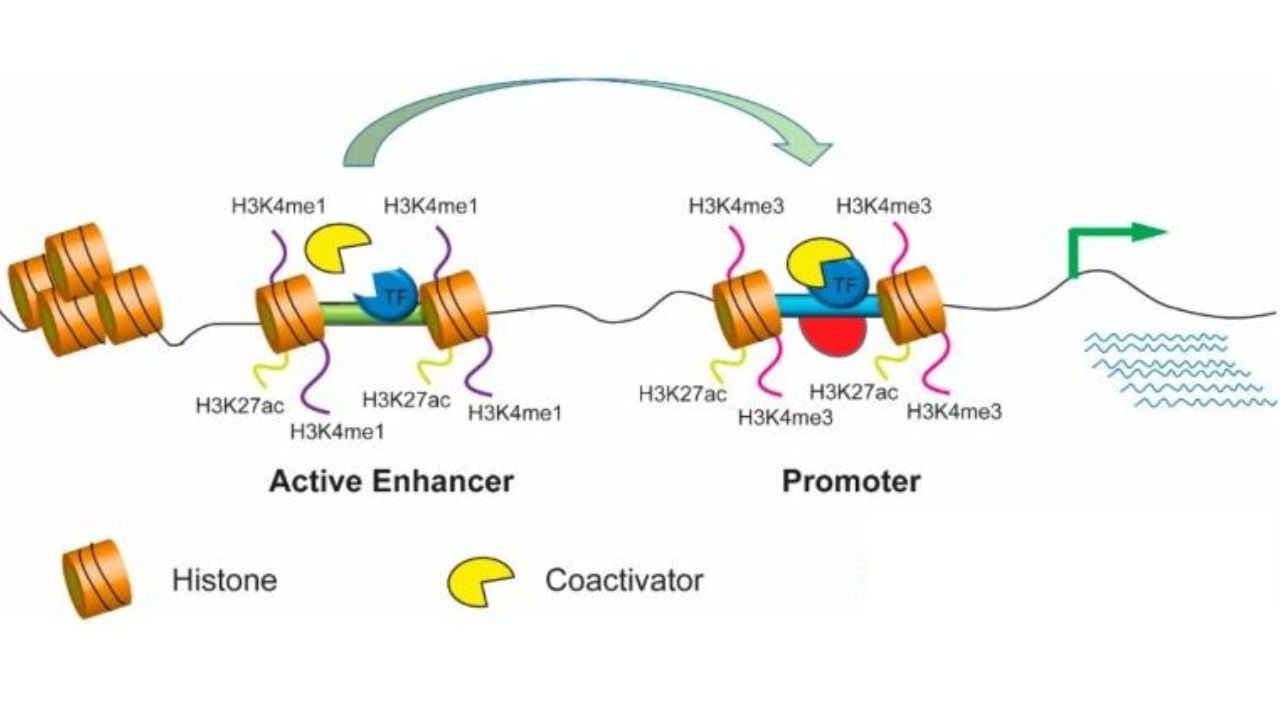 Epigenetic markers of enhancers and promoters. Cells. 2019.
Epigenetic markers of enhancers and promoters. Cells. 2019.
Cis-elements involved in transcriptional regulation also include insulators and silencers. Insulators can block the regulatory effects of certain cis-elements on gene expression and prevent heterochromatin diffusion; silencers inhibit transcription within a specific region.
Eukaryotic transcription initiation also involves three steps: first, recognition of the promoter to form a closed preinitiation complex; second, DNA unwinding to form an open complex; and finally, transition to the elongation phase via “promoter release.”
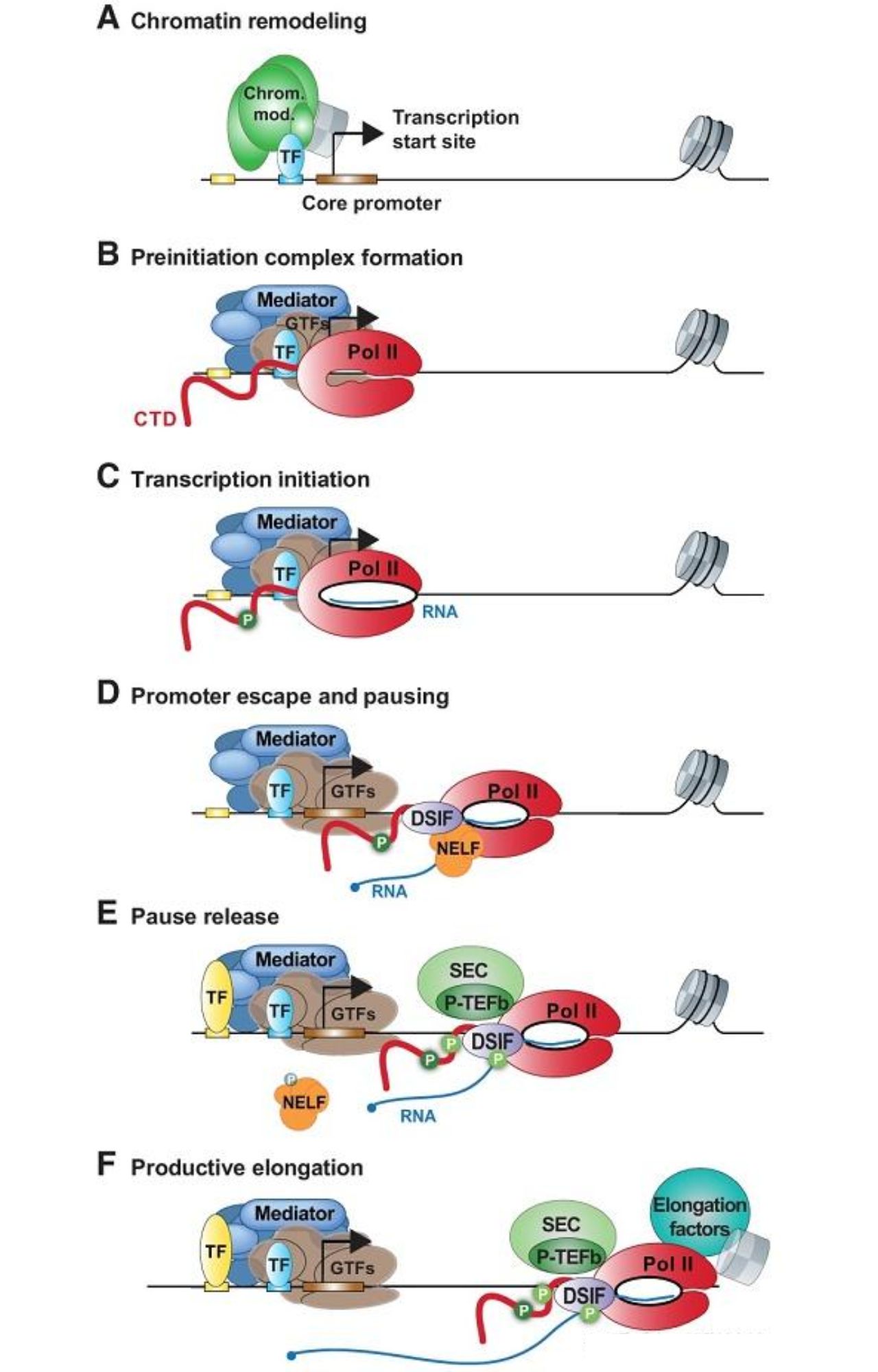 Early steps of eukaryotic transcription. Genes Dev. 2019.
Early steps of eukaryotic transcription. Genes Dev. 2019.
The initial closing complex (RPC) in prokaryotes consists of RNAP and a promoter, while promoter recognition in eukaryotes requires many transcription factors (TFs), which act similarly to σ factors. The initial closing complex in eukaryotes is called the preinitiation complex (PIC), which includes the mediator complex and various universal transcription factors (GTFs), such as TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH.
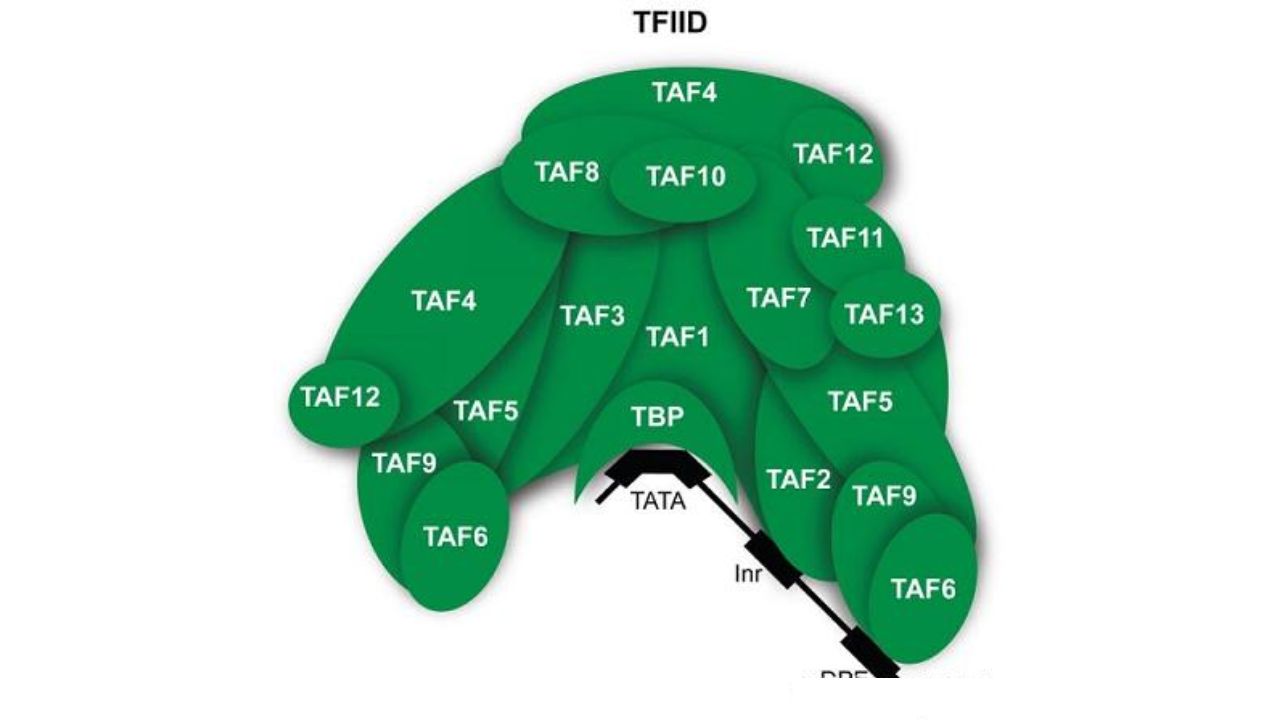 TFIID recognizes the core promoter. Nat Rev Genet. 2010
TFIID recognizes the core promoter. Nat Rev Genet. 2010
TFIID is a core promoter recognition factor for eukaryotic mRNA genes, a large assembly with a molecular weight greater than 1 MD. It includes TATA-binding protein (TBP) and 13-14 different TBP-associated factors (TAFs). TFIID recognizes the core promoter region on DNA, subsequently recruiting Pol II, TFIIH, and the Mediator complex, the pre-assembly initiation complex.
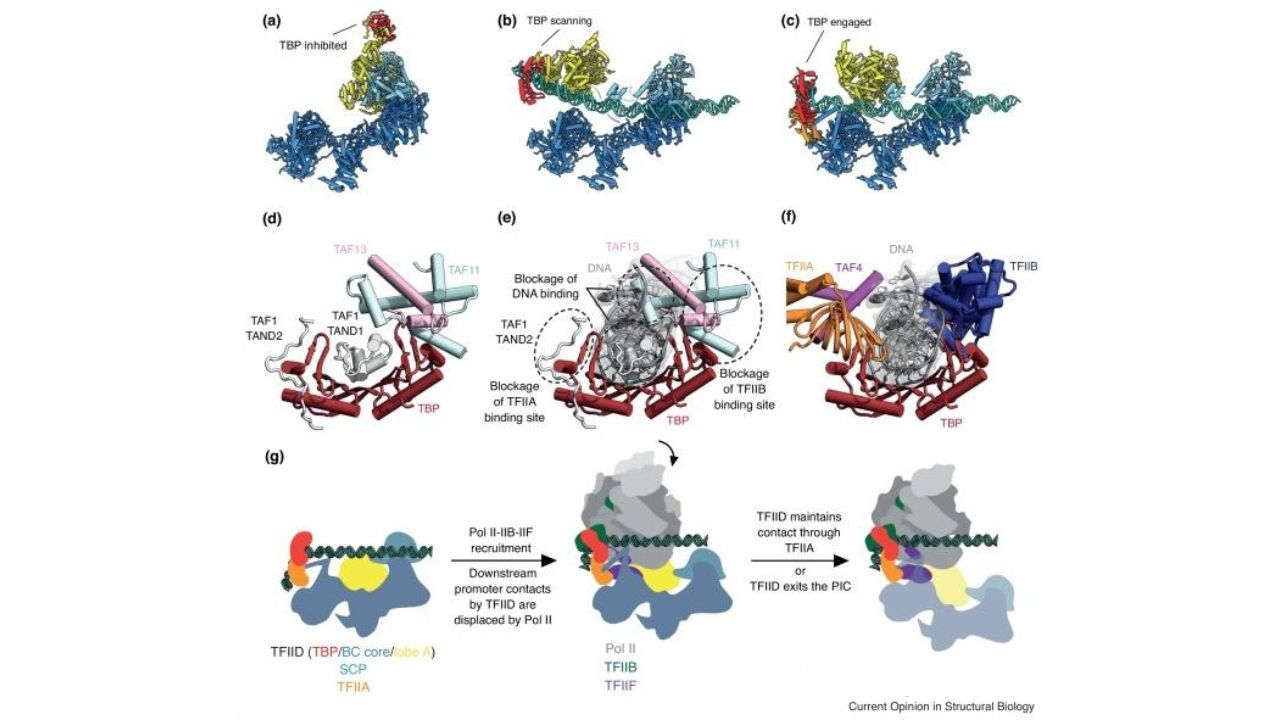 TFIID structure and RNAP recruitment. Curr Opin Struct Biol. 2019
TFIID structure and RNAP recruitment. Curr Opin Struct Biol. 2019
The Mediator is also a large complex with a molecular weight of several mega-molecules, involved in the recruitment and assembly of PICs. It interacts with the C-terminal domain (CTD) of the large subunit Rpb1 of Pol II; they bind when the CTD is unphosphorylated and dissociate upon phosphorylation. The Mediator also participates in transcriptional regulation, promoting or inhibiting transcription depending on its components.
After TFIIH is recruited, TFIIH-associated helicases promote the formation of transcription vesicles and allow template DNA to enter the Pol II active site. This initiates RNA synthesis, producing a hybrid double strand within the Pol II active site.
The RNA elongates, but the complex remains bound to the promoter, preventing the polymerase from moving forward and thus accumulating stress. After RNA larger than 10 nt is formed, the upstream region of the transcription bubble collapses, allowing the template and coding DNA strands to recombine.
It is thought that the energy gained in this process can propel the polymerase forward, enabling the promoter unwinding step. Of course, promoter unwinding is a complex process involving phosphorylation of the Pol II carboxy-terminal domain (CTD). CTD phosphorylation can interfere with its binding to the mediator, facilitating unwinding from the promoter.
It has been increasingly discovered that various RNAs are involved in transcription initiation. For example, enhancers transcribe enhancer RNA (eRNA), which can stabilize chromatin loops and may also participate in transcriptional elongation.
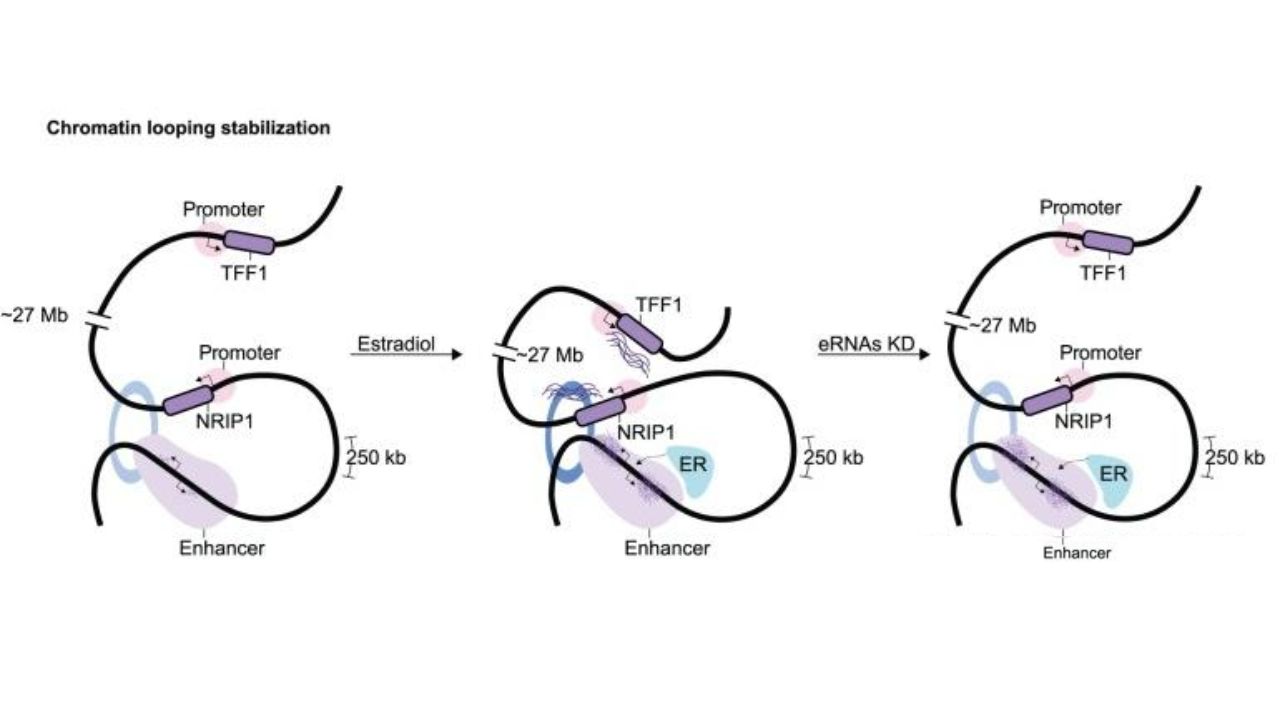 eRNA stabilizes chromatin loops. Epigenet Insights. 2019.
eRNA stabilizes chromatin loops. Epigenet Insights. 2019.
Some transcriptome studies have shown that the promoter regions of most genes transcribe non-coding RNAs, called promoter-associated noncoding RNAs (pancRNAs). These may promote or inhibit transcription through cis-elements, and several models exist (J Exp Clin Cancer Res. 2020).
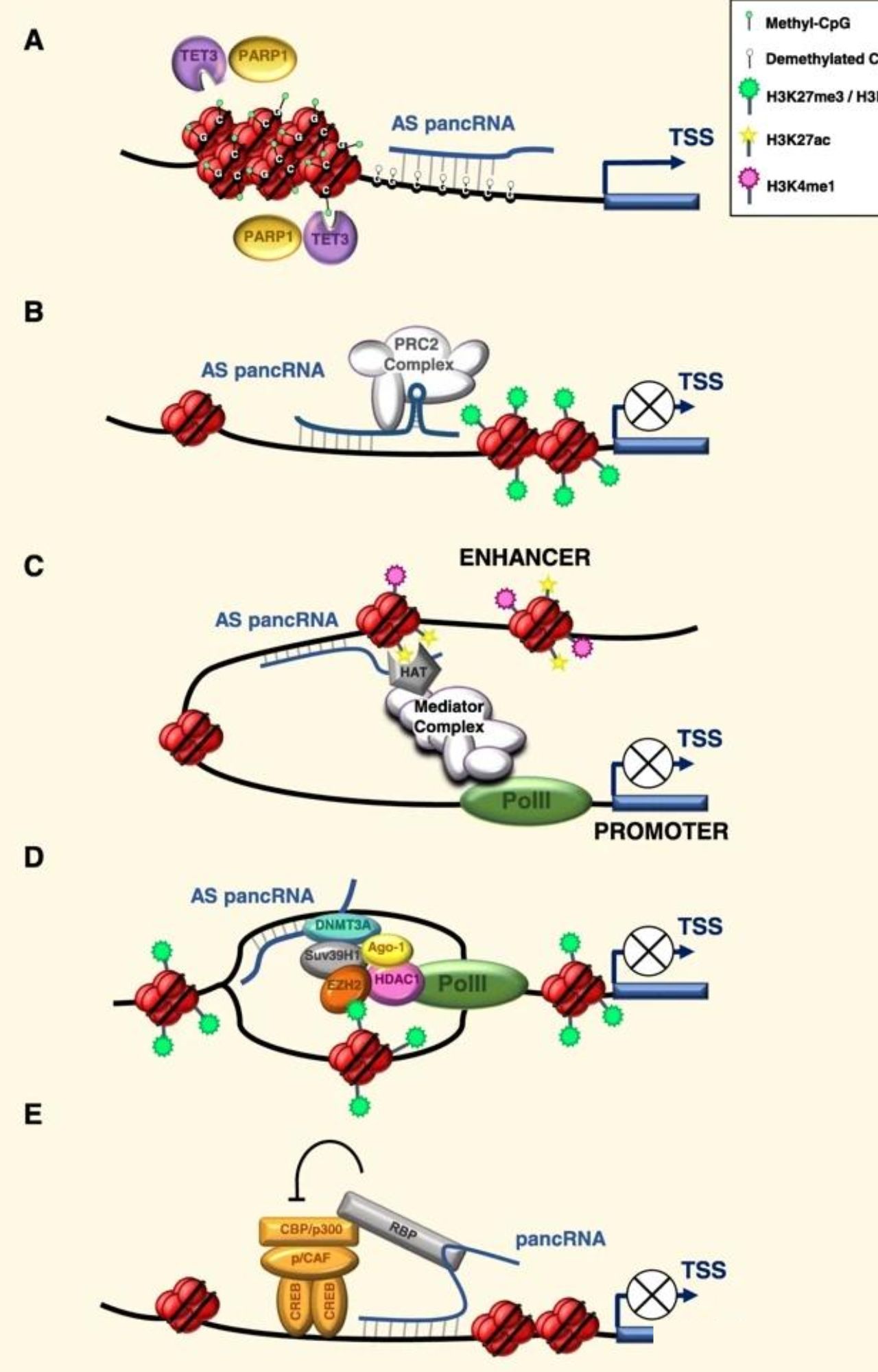 pancRNA mechanism model. J Exp Clin Cancer Res. 2020.
pancRNA mechanism model. J Exp Clin Cancer Res. 2020.
It was discovered in the 1960s that prokaryotic and eukaryotic RNA polymerases can initiate transcription in vitro using 2-8 nt RNA. In 2011, Goldman et al. found that 2-4 nt nanoRNAs in Pseudomonas aeruginosa can initiate transcription in vivo (Mol Cell. 2011).
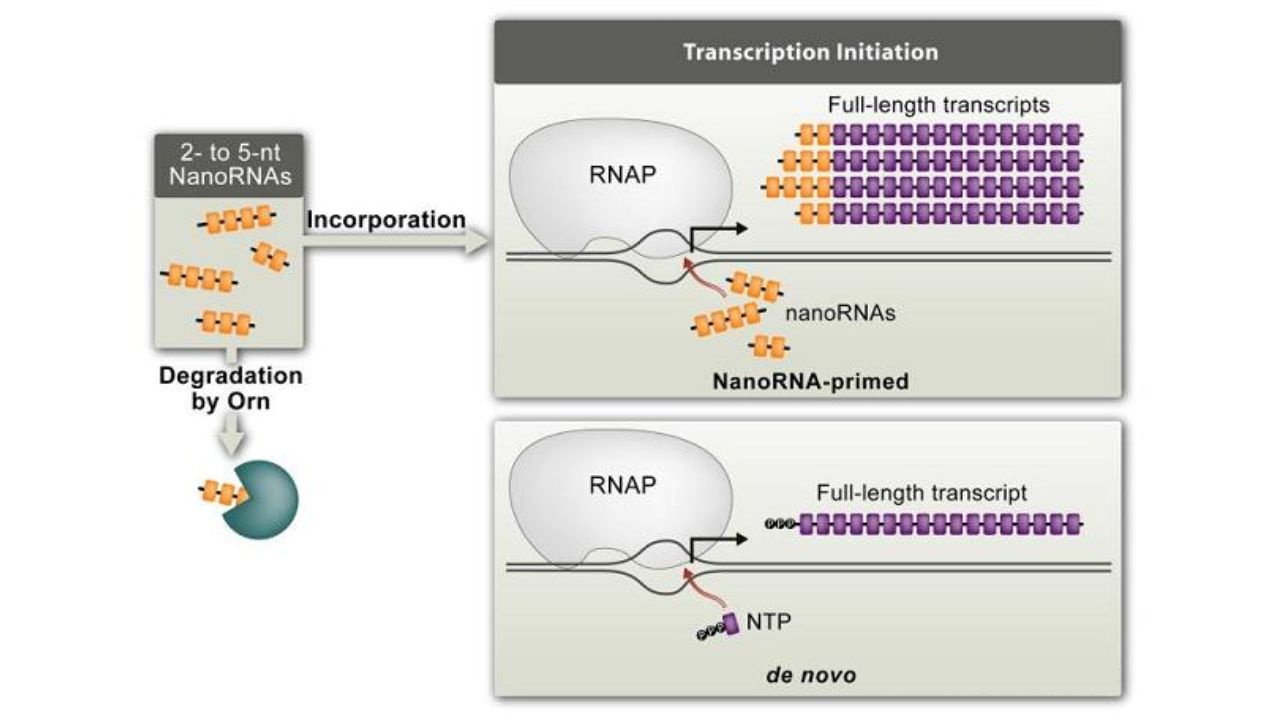 NanRNA regulates transcription initiation. J Mol Biol. 2011
NanRNA regulates transcription initiation. J Mol Biol. 2011
These extremely small RNAs can be degraded by specific oligonucleotides (Orn). Orn inactivation leads to the accumulation of nanoRNAs and triggers widespread RNA transcription. Therefore, nanoRNA-mediated initiation can serve as a mechanism for gene expression regulation.