What is Protein methylation?
Protein methylation refers to the enzymatic transfer of a methyl group to a specific residue in a protein, typically lysine or arginine, histidine, cysteine, or asparagine. Protein methylation is a common modification. It is often listed alongside acetylation because both are common epigenetic modifications and often occur on histones. However, from a chemical perspective, acetylation can be considered a short-chain fatty acylation modification, and this discussion focuses on methylation. The protein methylation donor is S-adenosylmethionine (SAM), and the acceptor is typically the ε-amino group of lysine and the guanidinium group of arginine. Methylation can also occur at the imidazole group of histidine, the amide groups of glutamine and asparagine, the sulfhydryl group of cysteine, the carboxyl group of cysteine, and the side chain carboxyl groups of glutamic acid and aspartic acid. For example, cysteine carboxyl methylation was mentioned in our discussion of protein prenylation. In some proteins with the CAAX sequence, the addition of the prenyl group cleaves the AAX residue, and the carboxyl group of cysteine is then methylated to form a methyl ester.
How does protein methylation occur?
Lysine residues can be mono-, di-, or tri-methylated. Arginine can be mono- or dimethylated, and dimethylation can be symmetrical or asymmetrical, depending on the methyltransferase. Humans express 27 lysine methyltransferases (KMTs) and 9 protein arginine methyltransferases (PRMTs). Lysine methylation was originally considered a permanent covalent mark, but it has since been discovered that lysine methylation can be transient and dynamically regulated by demethylation reactions. Many enzymes catalyze lysine demethylation, the largest of which are Jumonji C (JmjC) domain-containing demethylases, which are 2-oxoglutarate- and Fe2+-dependent dioxygenases. As a classic epigenetic modification, methylation of histones in nucleosomes is a crucial regulator of chromatin structure and gene transcriptional activity. Methylation at different sites can have different positive and negative effects on transcription. In addition to histones, a wide range of proteins can be methylated. Protein methylation can affect protein-protein interactions, protein-DNA or protein-RNA interactions, protein stability, subcellular localization, or enzymatic activity. Methylation modifications of many transcription factors can affect gene expression.
Differences between methylation, acetylation, and phosphorylation
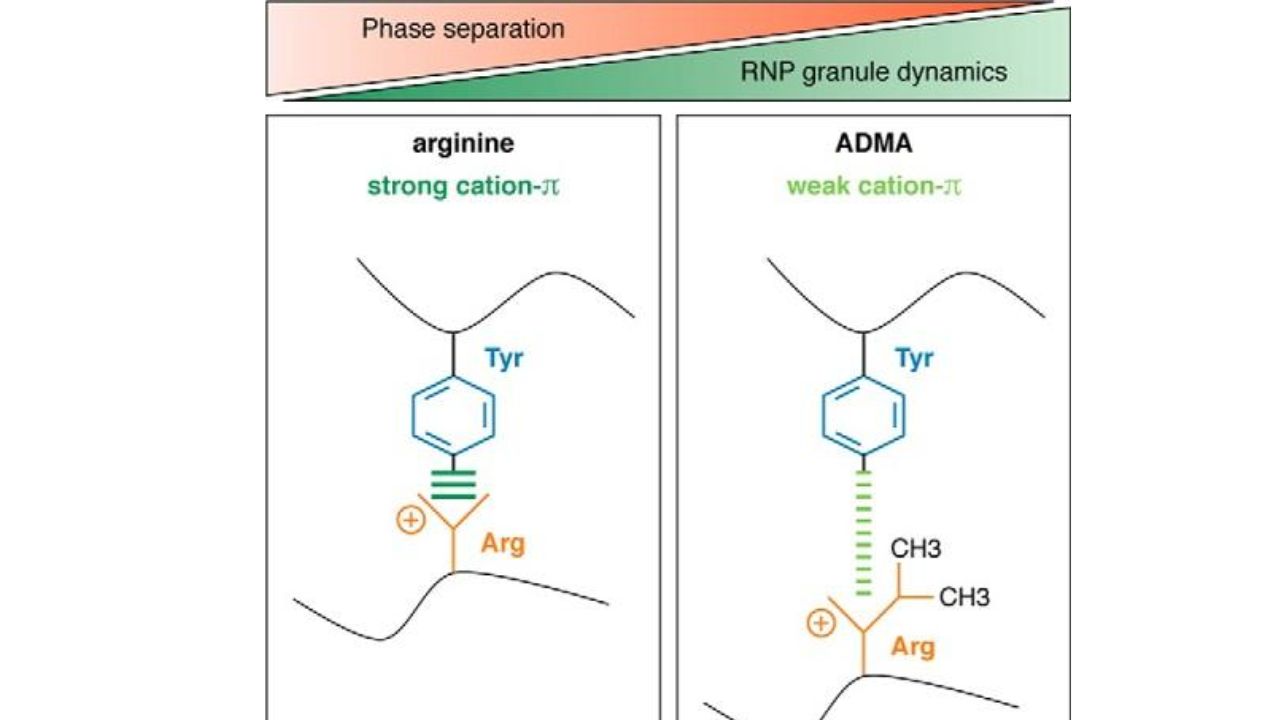
Unlike acetylation and phosphorylation, methylation does not affect the overall charge of a residue. However, the methyl group still influences its properties. A typical example is the effect of arginine methylation on phase separation. The positive charge of the arginine side chain attracts the π electrons of the aromatic ring, a type of cation-π interaction. This interaction often plays a crucial role in protein folding and protein-molecule interactions, such as in liquid-liquid phase separation (LLPS).
Condensates and droplets
Biomacromolecules form condensates, also known as droplets or membrane-less organelles (MLOs), through phase separation. These condensates are metastable and can solidify into viscous liquids, gels, or even solid amyloids. This process has been linked to a variety of physiological and pathological phenomena, including chromatin higher-order structure, gene expression, ribosome formation, DNA damage response, and neurodegenerative diseases. FUS (fused in sarcoma) is a multifunctional DNA/RNA binding protein that plays an important role in RNA transcription, splicing, transport, and translation. However, its dysfunction can lead to various neurodegenerative diseases, known as FUS proteinopathies, including FUS-related frontotemporal lobar degeneration/dementia (FTLD-FUS) and non-amyotrophic lateral sclerosis (ALS-FUS). The low-complexity (LC) structure of FUS can form antiparallel β-sheets, which stabilize its liquid droplet and gel states, thereby promoting certain phase separation processes, such as the formation of ribonucleoprotein (RNP) particles. Cation-π interactions between arginine and tyrosine are key drivers of this process. Studies have shown that methylation of arginine impairs this interaction, thereby reducing LLPS.
What are the consequences when methylation is inhibited?
RNP particles are involved in regulating local RNA and protein metabolism in subcellular locations, such as axon terminals and dendrites. Problems in this process can lead to diseases such as ALS and FTLD. It has been found that when methylation is inhibited, hypomethylated FUS (HYPO-FUS) is formed, which enhances LLPS and forms a large number of droplets and β-sheet-rich hydrogels, promoting amyloid deposition and ultimately leading to neurodegenerative diseases.